Scraping Drug Information from UpToDate.com
Curious to see the code for this project? Find it here on GitHub.
When I first applied to work as a Research Assistant, I was given the following coding challenge:
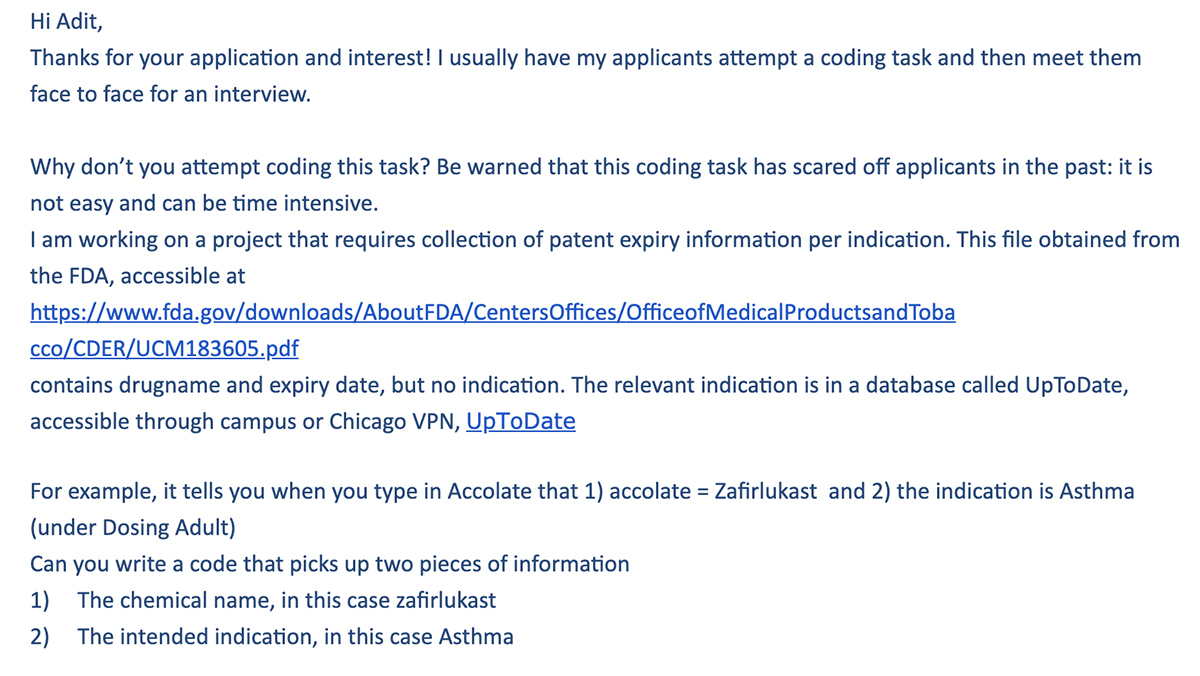
The FDA PDF linked in the picture above. And a link to UptoDate (note that you’ll need a subscription to access its drug information database — this was provided to me separately as a UChicago student).
Here’s an overview of the process I used to automate the drug information retrieval from UptoDate:
- I first converted the PDF to a plain text document. I used SCP to access the CSIL computers at UChicago, which run Linux. Linux has a nifty “pdftotext” feature, which outputs the formatted table into a reasonably well formatted text document.
- I used a Python script to select just the generic drug names from that text document (noticing that I could simply filter for Uppercase names).
- I then used Python with the Selenium Automated Google ChromeDriver on a macOS laptop to automatically login using my UChicago credentials, and then submit searches repeatedly on the UpToDate platform. Since I had a complete list of the generics’ names from Step 1, I used that for input on my searches.
At this point, I had search inputs (drug generic names) and access to the UpToDate search engine. The hardest part about the next step was that my script had to mimick the behavior of a real user. Imagine searching for “ALLEGRA” on Google. Eventually you’d find that the generic chemical name for Allegra is Fexofenadine, and symptoms indicating you should prescribe it are “Upper respiratory allergies: Chronic idiopathic urticaria”.
But to get that information, you’d probably have to click on a couple different websites, like WebMD, and scroll to the relevant information. Throughout that process, your brain actually does some simple tasks that would be complex for a computer (i.e. identifying that WebMD is a good link to click on, and scrolling to the part of the site with relevant information).
This was the challenge I had to enable my script to do.
- I noticed the database was based in AngularJS. To summarize, a consequence of this was that I couldn’t figure out how to write code that would “click on the first link.”
- I circumvented the issue by writing code that would click on a link with the text “: Drug Information” or “Systemic.” This worked 94% of the time (29/484) to bring me to the proper page to get the identification. I also used a similar process to get the chemical name, which worked 98.2% of the time.
- I similarly could not write code to identify which text on the page was the identification, since each drug info’s page was different in formatting. I tried my best to select for identification based off indentation under the heading “Dosing Adult”. I opted to include multiple indentications when a drug had more than just one. A bit of data cleaning may be required as a result for both chemical names and identifications.
Here’s what my script eventually produced!
Pretty nifty right? A process that could take a full day for almost 500 pharmaceutical drugs was performed in about an hour.
Although it took me about 4 days to come up with this automated solution 😅